1 Lab 1: Light
1.1 Getting the lab template
In RStudio, you’ll find the R Markdown template that you will update with your own results by following along with the GIF below:
Figure 1.1: Getting the Light R Markdown template
I’ve also created a new folder called Lab1-Light in my CHEM101_ChesterIsmay folder and named the resulting R Markdown file that was loaded in from the R Markdown template in chemistr the same thing: Lab1-Light.Rmd. You are highly encouraged to follow this same convention for all of your labs. (Note that as I mentioned earlier in this manual I do not have any spaces in the folder names or in the names of files here.) This will allow you to remember exactly where you stored the files and allow Danielle and I to easily find your files on the RStudio Server. Note that this folder was also created in the CHEM101_ChesterIsmay folder where the Rproj file resides. If you put your lab in the Home directory instead, Danielle and I won’t be able to access it since Home is the parent directory of CHEM101_YourName.
1.2 The parts of the lab template
1.2.1 The YAML header
I’ve already entered some values into my R Markdown file. First I gave a title to the R Markdown file: “Lab 1 - Light”. I then entered my name and the current date. All of these are in quotation marks since they all correspond to character strings and not numbers. It will likely still work if you don’t put quotation marks around the entries here, but it is recommended that you use the quotation marks to help you understand the use of quotation marks when you begin working with R code. (R won’t be as nice as YAML is on forgetting quotation marks when they are needed.)
This portion of the R Markdown file is called the YAML header. It stores metadata about your document. It is separated by --- before and --- after on their own lines. RStudio will warn you by coloring the --- red if you accidentally add a space before them.
You’ll need to make sure to include a space after each of the YAML entries. You’ll receive an error if you have something like that below, for example:
title: "Light Lab"
author: "Chester Ismay"
date:"August 30, 2016"
output: chemistr::chem_lab_wordThere needs to be a space after the : for the date field.
The last line here corresponds to the type of output that will be produced. In this case, I’ve created a nice way for you to output this R Markdown file to a DOCX (Word document) format. The chem_lab_word document format in chemistr creates a Word document with figure captions enabled by default and images with size 3 inches in height and 6 inches in width by default.
1.2.2 Initial R Chunk
On line 8 of the Light R Markdown template, you’ll see three backticks starting the line. This tells R Markdown to expect a block/chunk of code. This code is R code by the next designation {r. Next we give this R chunk a name setup and then specify chunk options immediately after the name and a comma. Here we have specified include=FALSE. We then close the specifications for the chunk with a }. This include=FALSE chunk option will run the R code when we press Knit button near the top of the code editor will not put the R code or its output into the document. We’ll press the Knit button in a bit to understand what it accomplishes.
You’ll see this setup chunk in all of the lab templates. It’s good practice to load any package you’ll need for your analysis at the top in a chunk like this. You may not initially understand why chemistr is needed but it will be needed when we create a table using the chem_table function, which resides in the chemistr package, in the table chunk beginning on line 21.
1.2.3 Headers
Immediately below this first R chunk is two hash tags. This corresponds to creating a header in our resulting document. The font immediately following the hash-tags will be larger and act as a divider in our document. The number of hash-tags you enter corresponds to how large you’d like to make the header. One hash-tag is the largest and six is the smallest. It is recommended that you use between one and three hash-tags for headers.
1.2.4 Enter data chunk
If you press the Knit button at this moment, you’ll receive an error. Let’s look into what this error means:
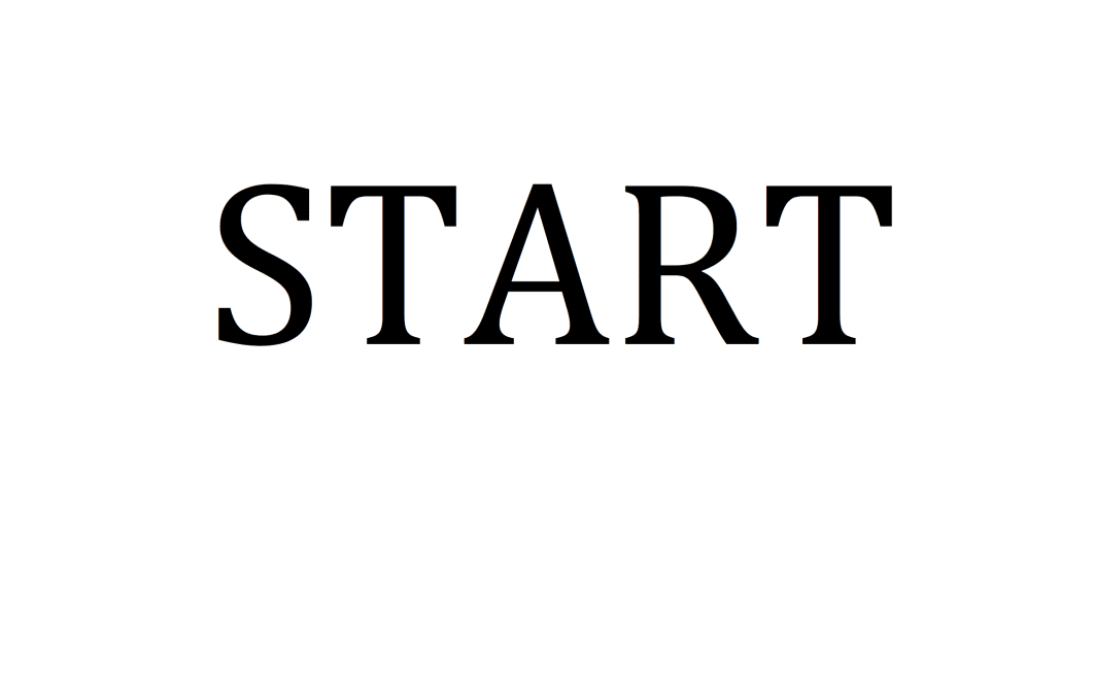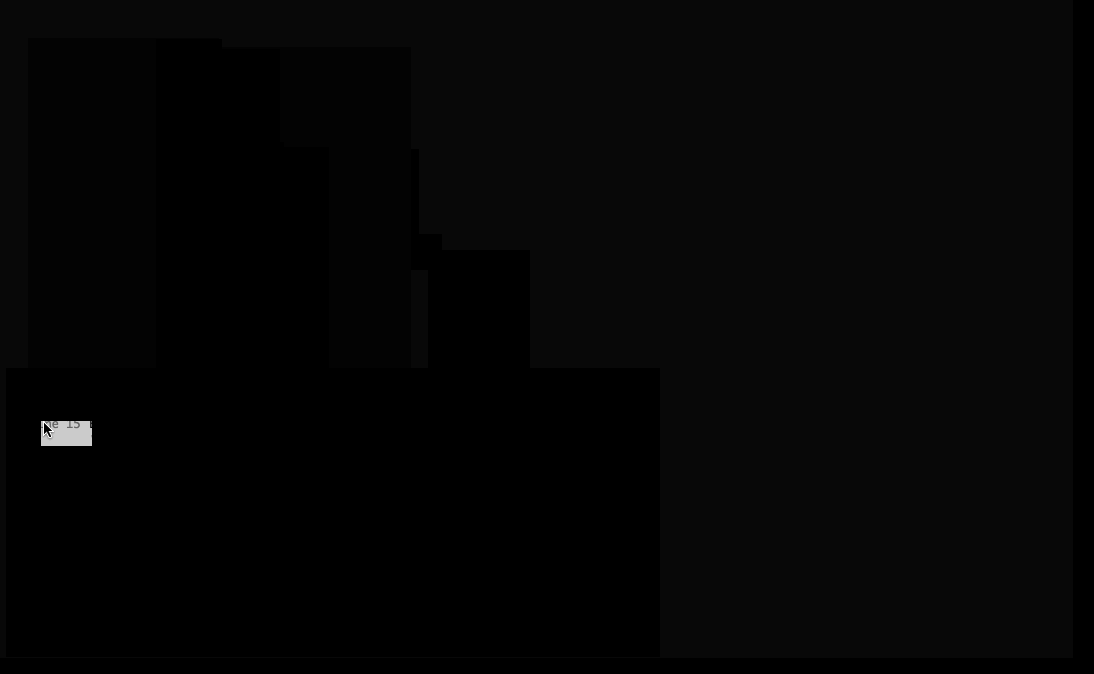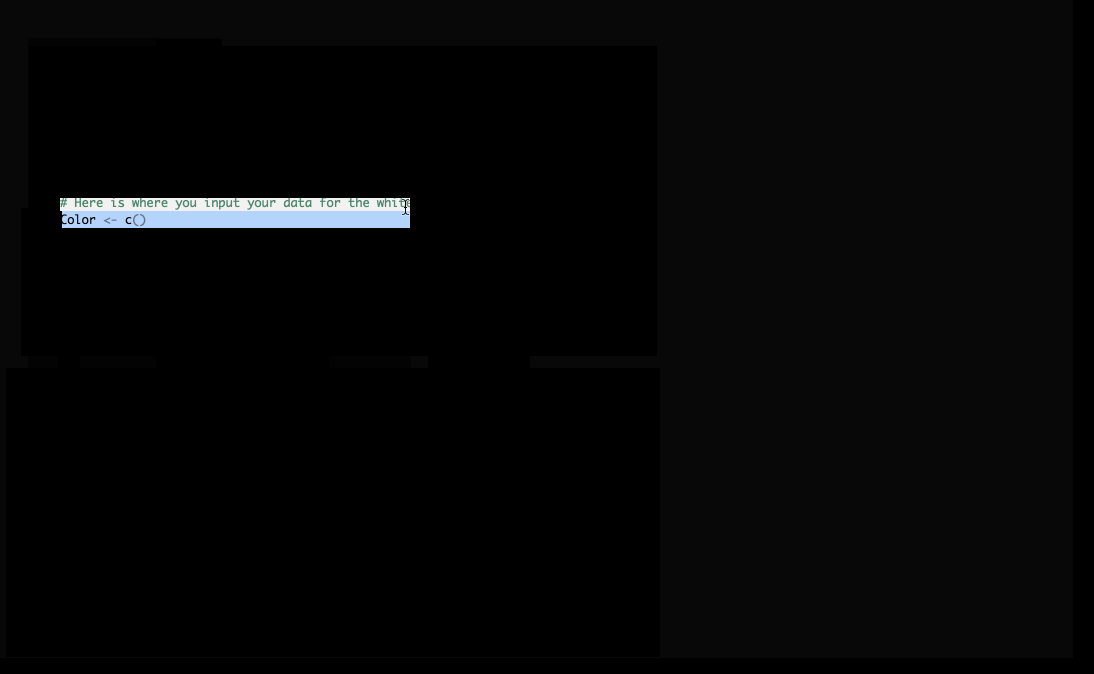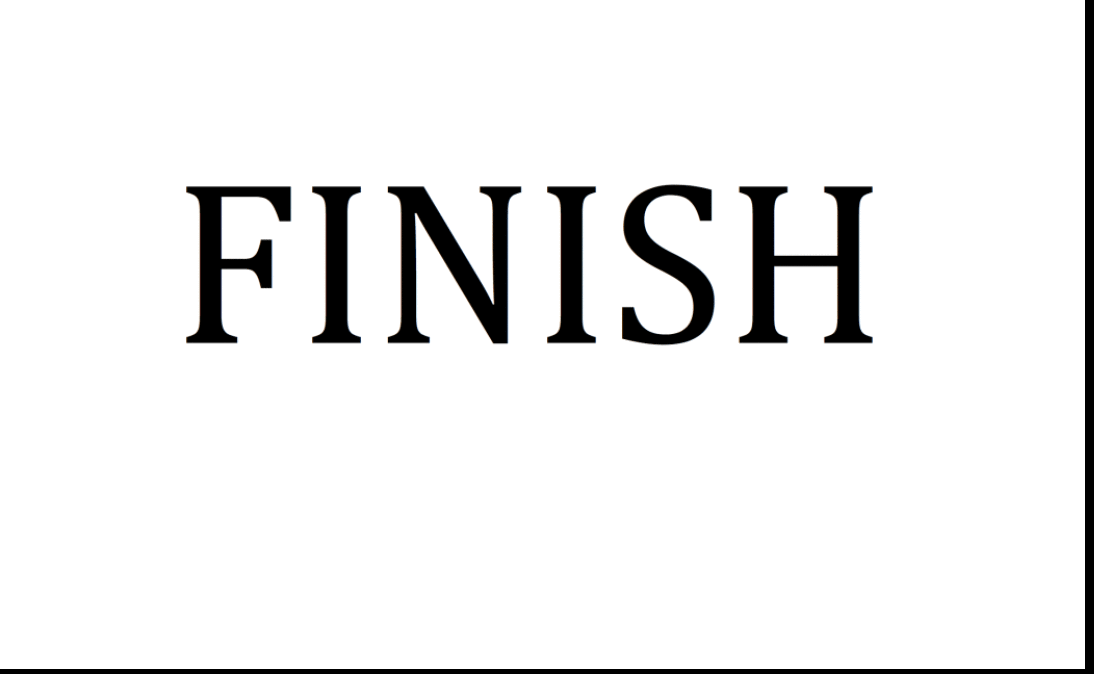
Figure 1.2: Initial Knit Error
Danielle has created lab templates that might not necessarily build without errors immediately for you. You should carefully read over the directions in each chunk (The comments after the # in the R chunks.) before pressing Knit. Here we see that the error occurs near line 15 of our R Markdown file. The Color and WavelengthRange vectors need to have some values entered. These results will correspond to what you observed in lab. I will enter some demo results in here and also show you how to use the R Console to check your results. You’ll see that the Color and WavelengthRange vectors must be of the same length when we create a data_frame to combine them together into “spreadsheet” format in the variable named Q1_data:

Figure 1.3: Enter values for Color and WavelengthRange
You should use the R Console to check your results in the R chunks by either entering the R code line by line into the Console or by pressing the Run Current Chunk button (the green Play button). We see here that Q1_data is 6 rows by 2 columns and stores our results in “spreadsheet” form as a data_frame.
Note that you’ll also receive an error if you have a different number of entries for WavelengthRange and Color. Data frames must have an equal number of rows when you combine the columns together via data_frame.
The chemistr package also loads the dplyr package where the data_frame function resides. If you are running the chunks by pressing the green “play” button, which sends the current chunk into the R console, you’ll need to remember to load all previous chunks as well. If you receive an error message such as
Error: could not find function "data_frame"this means that you have not loaded the chemistr package by sending the library(chemistr) code into the Console BEFORE you run the declarations for Color, Wavelength, and Q1_data <- data_frame(Color, WavelengthRange). The code chunks read like a book so if you are receiving errors of the “could not find” variety, it probably means you haven’t run all chunks up to the point of your current chunk in the Console. These sorts of errors are also referenced in Chapter 6 of “Getting Used to R, RStudio, and R Markdown”(???).
1.2.5 A nice table R chunk
The chem_table function will result in a nicely formatted table being produced in your resulting Word document. You’ll need to specify a caption inside the parentheses instead of the "A lovely caption for the table" default. You’ll also see that RStudio will create a second quotation mark when you might not want one. Carefully look over your document if you have errors and make sure there aren’t two quotation marks when you only wanted one. You’ll see this below:

Figure 1.4: Two quotation marks error
After fixing our error, we have now created a DOCX file that will have our nice table showing the Q1_data. Feel free to open up this Lab1_Light.docx file now and see that the table was created with the caption “Light Source Table”. You should probably enter a different and more specific caption instead though.
If you scroll down the R Markdown file, you’ll see that there are many more spots where we will be inserting pictures. R Markdown has nicely skipped over these chunks when we pressed Knit instead of halting and giving lots of confusing errors and we see that in the R Console window under the R Markdown tab.
1.3 Uploading and inserting external pictures
You’ll frequently be asked to add pictures into your lab reports that weren’t necessarily created using R. (We will see how to include plots created inside R in later labs though.) If you look over Q2 in the lab it is asking you to include a picture of the “Absorbance spectrum of water in a 1 cm cuvette”. Note that this is specified as the figure caption here for the first R chunk after Q2.
Mathematical text is also included by surrounding the mathematical expression with dollar signs. This is using LaTeX to produce nicely formatted math text. Here we see the expression \(H_{2}O\) is given using $H_{2}O$.
In order to include a picture, you’ll need to first upload it to the RStudio Server into the directory where the R Markdown file is located. The GIF below shows you how to do so. Remember that you’ll need to know where you saved your image on your computer to specify it here. This example GIF is recorded on a Mac but similar procedures should be done on a Linux or Windows machine.

Figure 1.5: Uploading a picture
The Water.jpg file has been uploaded to my Lab1-Light folder on the RStudio Server. The include_graphics function requires that you add the name of the file and its extension here. We now press the Knit button to see the resulting Word document:

Figure 1.6: Included photo and table in Word document
1.3.1 The remainder of the lab
In the remaining portions of the lab you’ll be asked to include other pictures from your lab sessions and also include a discussion of your results. This discussion will proceed in much the same way as you would add a discussion into a Word document or Google Doc. You’ll be able to immediately discuss what occurred in your R chunks right below them. This allows for the reader to easily follow your work.
1.4 Note on white space
As you look over the R Markdown document you’ll see that there is always a new line of white space between the discussion and the R chunks and also between each of the R chunks. It is highly recommended that you also follow this workflow. You’ll receive some strange errors at times if you try to stack everything together and it’s also much harder to follow for another reader of your document if you have everything bunched together. White space is your friend!
1.5 Spell-check
Just as I’m sure your English teachers have told you to spell check your documents before submitting, you are also encouraged to do so here. There is a built-in spell check option found near the Knit button.


Please run this and carefully read over your lab report before converting it to a PDF and submitting it to Moodle.
1.6 Converting your Word document to PDF
The directions for each lab on Moodle say to upload a PDF version of your lab. You’ll see how to create this PDF from inside Microsoft Word for Mac. A similar procedure can be done using LibreOffice (https://www.libreoffice.org/download/libreoffice-fresh/) on Linux, Mac, or Windows machines or Microsoft Word on a PC. You may also have the option to Save As a PDF there and you can get to this option by going to File -> Save As -> File Format: -> PDF on the Mac if you prefer.

Figure 1.7: Save as PDF on Mac from Microsoft Word