class: center, middle, inverse, title-slide # Statistical Inference: A Tidy Approach ### Dr. Chester Ismay <br> Senior Curriculum Lead at <a href="https://www.datacamp.com/">DataCamp</a> <br><br> <a href="http://github.com/ismayc"><i class="fa fa-github fa-fw"></i> ismayc</a><br> <a href="http://twitter.com/old_man_chester"> <i class="fa fa-twitter fa-fw"></i> <span class="citation">@old_man_chester</span></a><br> <a href="mailto:chester@datacamp.com"><i class="fa fa-paper-plane fa-fw"></i> chester@datacamp.com</a><br> ### 2018-04-13 <br>New England Statistical Symposium - UMass Amherst<br><br> Slides available at <a href="http://bit.ly/ness-infer" class="uri">http://bit.ly/ness-infer</a> <br> PDF slides at <a href="http://bit.ly/ness-infer-pdf" class="uri">http://bit.ly/ness-infer-pdf</a> --- class: inverse name: toc # Table of Contents Part 1 - [Data Wrangling](#wrangling) - [Data Visualization](#viz) - [Data Tidying](#tidying) Part 2 - [Resampling](#resampling) - [Inference](#inference) .footer[R code from throughout the slides is saved as an R script [here](https://raw.githubusercontent.com/ismayc/talks/master/ness-infer/slide_code.R)] --- layout: true class: inverse .footer[Slides available at http://bit.ly/ness-infer       [Return to Table of Contents](#toc)] --- ## Prior Installation Make sure you have the (current) up-to-date R, RStudio, and R packages - [Beginner's Guide](http://moderndive.com/2-getting-started.html) on ModernDive.com *** - [R (version 3.4.4)](https://cran.r-project.org/) - [RStudio (version 1.1.442)](https://www.rstudio.com/products/rstudio/download3/) - Run this in the RStudio Console ```r pkgs <- c("tidyverse", "moderndive", "gapminder", "nycflights13", "fivethirtyeight", "janitor", "ggplot2movies", "remotes") install.packages(pkgs) remotes::install_github("andrewpbray/infer") ``` --- ## Freely available information <center> <a href="http://moderndive.com"> <img src="img/wide_format.png" style = "width: 650px;"/> </a> </center> <center><h2>A Modern Dive into Data with R</h2></center> * Webpage: <http://moderndive.com> * Developmental version at <https://moderndive.netlify.com> * [GitHub Repo](https://github.com/moderndive/moderndiver-book) * Please [signup](http://eepurl.com/cBkItf) for our mailing list! --- layout: false class: center, middle <img src="img/flowchart.png" style="width: 750px"/> --- layout: true class: inverse .footer[Slides available at http://bit.ly/ness-infer       [Return to Table of Contents](#toc)] --- ## Designed for the novice / Nice for the practioner <center> <a href="http://moderndive.netlify.com/2-getting-started.html"> <img src="img/engine.png" style = "width: 600px;"/> </a> </center> --- ## Designed for the novice / Nice for the practioner <center> <a href="http://moderndive.netlify.com/2-getting-started.html"> <img src="img/appstore.png" style = "width: 620px;"/> </a> </center> --- class: normal, center, middle # R Data Types --- ## The bare minimum needed for understanding today Vector/variable - Type of vector (`int`, `num` or `dbl`, `chr`, `lgl`, `date`) -- Data frame - Vectors of (potentially) different types - Each vector has the same number of rows --- ## The bare minimum needed for understanding today ```r library(tibble) library(lubridate) ex1 <- data_frame( vec1 = c(1980, 1990, 2000, 2010), vec2 = c(1L, 2L, 3L, 4L), vec3 = c("low", "low", "high", "high"), vec4 = c(TRUE, FALSE, FALSE, FALSE), vec5 = ymd(c("2017-05-23", "1776/07/04", "1983-05/31", "1908/04-01")) ) ex1 ``` -- ``` # A tibble: 4 x 5 vec1 vec2 vec3 vec4 vec5 <dbl> <int> <chr> <lgl> <date> 1 1980. 1 low TRUE 2017-05-23 2 1990. 2 low FALSE 1776-07-04 3 2000. 3 high FALSE 1983-05-31 4 2010. 4 high FALSE 1908-04-01 ``` --- class: center, middle # Welcome to the [tidyverse](https://blog.rstudio.org/2016/09/15/tidyverse-1-0-0/)! The `tidyverse` is a collection of R packages that share common philosophies and are designed to work together. <br><br> <a href="http://tidyverse.tidyverse.org/logo.png"><img src="figure/tidyverse.png" style="width: 200px;"/></a> --- # First motivating example for today <a href="http://gitsense.github.io/images/wealth.gif"><img src="figure/wealth.gif" style="width: 700px;"/></a> - Inspired by the late, great [Hans Rosling](https://www.youtube.com/watch?v=jbkSRLYSojo) --- layout: false class: center, middle name: wrangling # Data Wrangling <center> <a href="https://dplyr.tidyverse.org"> <img src="img/dplyr_hex.png" style = "width: 400px;"/> </a> </center> --- layout: true class: inverse .footer[Slides available at http://bit.ly/ness-infer       [Return to Table of Contents](#toc)] --- ## The [`gapminder` package](https://github.com/jennybc/gapminder) ```r library(gapminder) gapminder ``` ``` # A tibble: 1,704 x 6 country continent year lifeExp pop gdpPercap <fct> <fct> <int> <dbl> <int> <dbl> 1 Afghanistan Asia 1952 28.8 8425333 779. 2 Afghanistan Asia 1957 30.3 9240934 821. 3 Afghanistan Asia 1962 32.0 10267083 853. 4 Afghanistan Asia 1967 34.0 11537966 836. 5 Afghanistan Asia 1972 36.1 13079460 740. 6 Afghanistan Asia 1977 38.4 14880372 786. 7 Afghanistan Asia 1982 39.9 12881816 978. 8 Afghanistan Asia 1987 40.8 13867957 852. 9 Afghanistan Asia 1992 41.7 16317921 649. 10 Afghanistan Asia 1997 41.8 22227415 635. # ... with 1,694 more rows ``` --- ## Base R versus the `tidyverse` - The mean life expectancy across all years for Asia -- ```r # Base R asia <- gapminder[gapminder$continent == "Asia", ] mean(asia$lifeExp) ``` ``` [1] 60.065 ``` -- ```r library(dplyr) gapminder %>% filter(continent == "Asia") %>% summarize(mean_exp = mean(lifeExp)) ``` ``` [1] 60.065 ``` --- ## The pipe `%>%` <img src="figure/pipe.png" style="width: 240px;"/>       <img src="figure/MagrittePipe.jpg" style="width: 300px;"/> -- - A way to chain together commands - Can be read as "and then" when reading over code -- ```r library(dplyr) gapminder %>% filter(continent == "Asia") %>% summarize(mean_exp = mean(lifeExp)) ``` --- name: fivemv # [The Five Main Verbs (5MV)](http://moderndive.com/5-wrangling.html) of data wrangling - [`filter()`](#filter) - [`summarize()`](#summarize) - [`group_by()`](#groupby) - [`mutate()`](#mutate) - [`arrange()`](#arrange) --- name: filter ## `filter()` - Select a subset of the rows of a data frame. - Arguments are "filters" that you'd like to apply. -- ```r library(gapminder); library(dplyr) gap_2007 <- gapminder %>% filter(year == 2007) head(gap_2007, 4) ``` ``` # A tibble: 4 x 6 country continent year lifeExp pop gdpPercap <fct> <fct> <int> <dbl> <int> <dbl> 1 Afghanistan Asia 2007 43.8 31889923 975. 2 Albania Europe 2007 76.4 3600523 5937. 3 Algeria Africa 2007 72.3 33333216 6223. 4 Angola Africa 2007 42.7 12420476 4797. ``` - Use `==` to compare a variable to a value --- ## Logical operators - Use `|` to check for any in multiple filters being true: -- ```r gapminder %>% filter(year == 2002 | continent == "Asia") %>% sample_n(8) ``` -- ``` # A tibble: 8 x 6 country continent year lifeExp pop gdpPercap <fct> <fct> <int> <dbl> <int> <dbl> 1 Iraq Asia 1997 58.8 20775703 3076. 2 Korea, Rep. Asia 1997 74.6 46173816 15994. 3 Bangladesh Asia 1952 37.5 46886859 684. 4 France Europe 2002 79.6 59925035 28926. 5 Kuwait Asia 1957 58.0 212846 113523. 6 Iran Asia 1962 49.3 22874000 4187. 7 Myanmar Asia 1982 58.1 34680442 424. 8 Canada Americas 2002 79.8 31902268 33329. ``` --- ## Logical operators - Use `,` to check for all of multiple filters being true: -- ```r gapminder %>% filter(year == 2002, continent == "Asia") ``` ``` # A tibble: 8 x 6 country continent year lifeExp pop gdpPercap <fct> <fct> <int> <dbl> <int> <dbl> 1 Afghanistan Asia 2002 42.1 25268405 727. 2 Bahrain Asia 2002 74.8 656397 23404. 3 Bangladesh Asia 2002 62.0 135656790 1136. 4 Cambodia Asia 2002 56.8 12926707 896. 5 China Asia 2002 72.0 1280400000 3119. 6 Hong Kong, China Asia 2002 81.5 6762476 30209. 7 India Asia 2002 62.9 1034172547 1747. 8 Indonesia Asia 2002 68.6 211060000 2874. ``` --- ## Logical operators - Use `%in%` to check for any being true <br> (shortcut to using `|` repeatedly with `==`) -- ```r gapminder %>% filter(country %in% c("Argentina", "Belgium", "Mexico"), year %in% c(1987, 1992)) ``` -- ``` # A tibble: 6 x 6 country continent year lifeExp pop gdpPercap <fct> <fct> <int> <dbl> <int> <dbl> 1 Argentina Americas 1987 70.8 31620918 9140. 2 Argentina Americas 1992 71.9 33958947 9308. 3 Belgium Europe 1987 75.4 9870200 22526. 4 Belgium Europe 1992 76.5 10045622 25576. 5 Mexico Americas 1987 69.5 80122492 8688. 6 Mexico Americas 1992 71.5 88111030 9472. ``` --- name: summarize ## `summarize()` - Any numerical summary that you want to apply to a column of a data frame is specified within `summarize()`. ```r stats_1997 <- gapminder %>% filter(year == 1997) %>% summarize(max_exp = max(lifeExp), sd_exp = sd(lifeExp)) stats_1997 ``` -- ``` # A tibble: 1 x 2 max_exp sd_exp <dbl> <dbl> 1 80.7 11.6 ``` --- name: groupby ### Combining `summarize()` with `group_by()` When you'd like to determine a numerical summary for all levels of a different categorical variable ```r max_exp_1997_by_cont <- gapminder %>% filter(year == 1997) %>% group_by(continent) %>% summarize(max_exp = max(lifeExp), sd_exp = sd(lifeExp)) max_exp_1997_by_cont ``` -- ``` # A tibble: 5 x 3 continent max_exp sd_exp <fct> <dbl> <dbl> 1 Africa 74.8 9.10 2 Americas 78.6 4.89 3 Asia 80.7 8.09 4 Europe 79.4 3.10 5 Oceania 78.8 0.905 ``` --- name: mutate ## `mutate()` - Allows you to 1. <font color="yellow">create a new variable with a specific value</font> OR 2. create a new variable based on other variables OR 3. change the contents of an existing variable -- ```r gap_plus <- gapminder %>% mutate(just_one = 1) head(gap_plus, 4) ``` ``` # A tibble: 4 x 7 country continent year lifeExp pop gdpPercap just_one <fct> <fct> <int> <dbl> <int> <dbl> <dbl> 1 Afghanistan Asia 1952 28.8 8425333 779. 1. 2 Afghanistan Asia 1957 30.3 9240934 821. 1. 3 Afghanistan Asia 1962 32.0 10267083 853. 1. 4 Afghanistan Asia 1967 34.0 11537966 836. 1. ``` --- ## `mutate()` - Allows you to 1. create a new variable with a specific value OR 2. <font color="yellow">create a new variable based on other variables</font> OR 3. change the contents of an existing variable -- ```r gap_w_gdp <- gapminder %>% mutate(gdp = pop * gdpPercap) sample_n(gap_w_gdp, 4) ``` ``` # A tibble: 4 x 7 country continent year lifeExp pop gdpPercap gdp <fct> <fct> <int> <dbl> <int> <dbl> <dbl> 1 Comoros Africa 1967 46.5 217378 1876. 407807572. 2 Paraguay Americas 1992 68.2 4483945 4196. 18816476471. 3 Mauritius Africa 1967 61.6 789309 2475. 1953845681. 4 Chile Americas 1997 75.8 14599929 10118. 147722858046. ``` --- ## `mutate()` - Allows you to 1. create a new variable with a specific value OR 2. create a new variable based on other variables OR 3. <font color="yellow">change the contents of an existing variable</font> -- ```r gap_weird <- gapminder %>% mutate(pop = pop + 1000) head(gap_weird, 4) ``` ``` # A tibble: 4 x 6 country continent year lifeExp pop gdpPercap <fct> <fct> <int> <dbl> <dbl> <dbl> 1 Afghanistan Asia 1952 28.8 8426333. 779. 2 Afghanistan Asia 1957 30.3 9241934. 821. 3 Afghanistan Asia 1962 32.0 10268083. 853. 4 Afghanistan Asia 1967 34.0 11538966. 836. ``` --- name: arrange ## `arrange()` - Reorders the rows in a data frame based on the values of one or more variables -- ```r gapminder %>% arrange(year, country) %>% head(10) ``` ``` # A tibble: 10 x 6 country continent year lifeExp pop gdpPercap <fct> <fct> <int> <dbl> <int> <dbl> 1 Afghanistan Asia 1952 28.8 8425333 779. 2 Albania Europe 1952 55.2 1282697 1601. 3 Algeria Africa 1952 43.1 9279525 2449. 4 Angola Africa 1952 30.0 4232095 3521. 5 Argentina Americas 1952 62.5 17876956 5911. 6 Australia Oceania 1952 69.1 8691212 10040. 7 Austria Europe 1952 66.8 6927772 6137. 8 Bahrain Asia 1952 50.9 120447 9867. 9 Bangladesh Asia 1952 37.5 46886859 684. 10 Belgium Europe 1952 68.0 8730405 8343. ``` --- ## `arrange()` - Can also put into descending order -- ```r gapminder %>% filter(year > 2000) %>% arrange(desc(lifeExp)) %>% head(10) ``` ``` # A tibble: 10 x 6 country continent year lifeExp pop gdpPercap <fct> <fct> <int> <dbl> <int> <dbl> 1 Japan Asia 2007 82.6 127467972 31656. 2 Hong Kong, China Asia 2007 82.2 6980412 39725. 3 Japan Asia 2002 82.0 127065841 28605. 4 Iceland Europe 2007 81.8 301931 36181. 5 Switzerland Europe 2007 81.7 7554661 37506. 6 Hong Kong, China Asia 2002 81.5 6762476 30209. 7 Australia Oceania 2007 81.2 20434176 34435. 8 Spain Europe 2007 80.9 40448191 28821. 9 Sweden Europe 2007 80.9 9031088 33860. 10 Israel Asia 2007 80.7 6426679 25523. ``` --- ## Don't mix up `arrange` and `group_by` - `group_by` is used (mostly) with `summarize` to calculate summaries over groups - `arrange` is used for sorting --- ## Don't mix up `arrange` and `group_by` This doesn't really do anything useful ```r gapminder %>% group_by(year) ``` ``` # A tibble: 1,704 x 6 # Groups: year [12] country continent year lifeExp pop gdpPercap <fct> <fct> <int> <dbl> <int> <dbl> 1 Afghanistan Asia 1952 28.8 8425333 779. 2 Afghanistan Asia 1957 30.3 9240934 821. 3 Afghanistan Asia 1962 32.0 10267083 853. 4 Afghanistan Asia 1967 34.0 11537966 836. 5 Afghanistan Asia 1972 36.1 13079460 740. 6 Afghanistan Asia 1977 38.4 14880372 786. 7 Afghanistan Asia 1982 39.9 12881816 978. 8 Afghanistan Asia 1987 40.8 13867957 852. 9 Afghanistan Asia 1992 41.7 16317921 649. 10 Afghanistan Asia 1997 41.8 22227415 635. # ... with 1,694 more rows ``` --- ## Don't mix up `arrange` and `group_by` But this does ```r gapminder %>% arrange(year) ``` ``` # A tibble: 1,704 x 6 country continent year lifeExp pop gdpPercap <fct> <fct> <int> <dbl> <int> <dbl> 1 Afghanistan Asia 1952 28.8 8425333 779. 2 Albania Europe 1952 55.2 1282697 1601. 3 Algeria Africa 1952 43.1 9279525 2449. 4 Angola Africa 1952 30.0 4232095 3521. 5 Argentina Americas 1952 62.5 17876956 5911. 6 Australia Oceania 1952 69.1 8691212 10040. 7 Austria Europe 1952 66.8 6927772 6137. 8 Bahrain Asia 1952 50.9 120447 9867. 9 Bangladesh Asia 1952 37.5 46886859 684. 10 Belgium Europe 1952 68.0 8730405 8343. # ... with 1,694 more rows ``` --- ## Practice Use the [5MV](#fivemv) to answer problems from R data packages, e.g., [`nycflights13::weather`] <!-- Lay out what the resulting table should look like on paper first. --> 1. What is the maximum arrival delay for each carrier departing JFK? [`nycflights13::flights`] 2. Calculate for each movie the domestic return on investment for 2013 scaled data descending by ROI <br> [`fivethirtyeight::bechdel`] --- layout: false class: center, middle name: viz # Data Visualization <center> <a href="https://ggplot2.tidyverse.org"> <img src="img/ggplot2_hex.png" style = "width: 400px;"/> </a> </center> --- layout: true class: inverse .footer[Slides available at http://bit.ly/ness-infer       [Return to Table of Contents](#toc)] --- <img src="slide_deck_files/figure-html/unnamed-chunk-29-1.png" style="display: block; margin: auto;" /> - What are the variables here? - What is the observational unit? - How are the variables mapped to aesthetics? --- class: center, middle ## Grammar of Graphics Wilkinson (2005) laid out the proposed <br> "Grammar of Graphics" <br> <a href="http://www.powells.com/book/the-grammar-of-graphics-9780387245447"><img src="figure/graphics.jpg" style="width: 200px;"></a> --- class: center, middle ## Grammar of Graphics in R Wickham implemented the grammar in R <br> in the `ggplot2` package <br> <a href="http://www.powells.com/book/ggplot2-elegant-graphics-for-data-analysis-9783319242750/68-428"><img src="figure/ggplot2.jpg" style="width: 200px;"></a> --- class: center, middle ## What is a statistical graphic? -- ## A `mapping` of <br> `data` variables -- ## to <br> `aes()`thetic attributes -- ## of <br> `geom_`etric objects. --- class: inverse, center, middle # Back to Basics --- ## Old school - Sketch the graphics below on paper, where the `x`-axis is variable `A` and the `y`-axis is variable `B` ``` # A tibble: 4 x 4 A B C D <dbl> <dbl> <dbl> <chr> 1 1980. 1. 3. cold 2 1990. 2. 2. cold 3 2000. 3. 1. hot 4 2010. 4. 2. hot ``` <!-- Copy to chalkboard/whiteboard --> 1. <small>A scatter plot</small> 1. <small>A scatter plot where the `color` of the points corresponds to `D`</small> 1. <small>A scatter plot where the `size` of the points corresponds to `C`</small> --- layout: true class: inverse .footer[Slides available at http://bit.ly/ness-infer       [Return to Table of Contents](#toc)] --- ## Reproducing the plots in `ggplot2` ### 1. A scatterplot ```r library(ggplot2) ggplot(data = simple_ex, mapping = aes(x = A, y = B)) + geom_point() ``` -- <!-- --> --- ## Reproducing the plots in `ggplot2` ### 2. A scatter plot where the `color` of the points corresponds to `D` ```r library(ggplot2) ggplot(data = simple_ex, mapping = aes(x = A, y = B)) + geom_point(mapping = aes(color = D)) ``` -- <!-- --> --- ## Reproducing the plots in `ggplot2` ### 3. A scatter plot where the `size` of the points corresponds to `C` ```r library(ggplot2) ggplot(data = simple_ex, mapping = aes(x = A, y = B, size = C)) + geom_point() ``` -- <!-- --> --- # [The Five-Named Graphs](http://moderndive.com/3-viz.html#FiveNG) ## The 5NG of data viz - Scatterplot: `geom_point()` - Line graph: `geom_line()` - Histogram: `geom_histogram()` - Boxplot: `geom_boxplot()` - Bar graph: `geom_bar()` --- class: center, middle ## More examples --- ## Histogram ```r library(nycflights13) ggplot(data = weather, mapping = aes(x = humid)) + geom_histogram(bins = 20, color = "black", fill = "darkorange") ``` <!-- --> --- ## Boxplot (broken) ```r library(nycflights13) ggplot(data = weather, mapping = aes(x = month, y = humid)) + geom_boxplot() ``` <!-- --> --- ## Boxplot (fixed) ```r library(nycflights13) ggplot(data = weather, mapping = aes(x = factor(month), y = humid)) + geom_boxplot() ``` <!-- --> --- ## Bar graph ```r library(fivethirtyeight) ggplot(data = bechdel, mapping = aes(x = clean_test)) + geom_bar() ``` <!-- --> --- ## How about over time? - One more variable to create with `dplyr` ```r library(dplyr) year_bins <- c("'70-'74", "'75-'79", "'80-'84", "'85-'89", "'90-'94", "'95-'99", "'00-'04", "'05-'09", "'10-'13") bechdel <- bechdel %>% mutate(five_year = cut(year, breaks = seq(1969, 2014, 5), labels = year_bins)) ``` --- ## How about over time? (Stacked) ```r library(fivethirtyeight) library(ggplot2) ggplot(data = bechdel, mapping = aes(x = five_year, fill = clean_test)) + geom_bar() + scale_fill_brewer(type = "qual") # set colors ``` <!-- --> --- ## How about over time? (Side-by-side) ```r library(fivethirtyeight) library(ggplot2) ggplot(data = bechdel, mapping = aes(x = five_year, fill = clean_test)) + geom_bar(position = "dodge") + scale_fill_brewer(type = "qual") ``` <!-- --> --- ## How about over time? (Stacked proportional) ```r library(fivethirtyeight) library(ggplot2) ggplot(data = bechdel, mapping = aes(x = five_year, fill = clean_test)) + geom_bar(position = "fill", color = "black") + scale_fill_brewer(type = "qual") ``` <!-- --> --- class: center, middle ## `ggplot2` is for beginners and for data science professionals! <a href="https://fivethirtyeight.com/features/the-dollar-and-cents-case-against-hollywoods-exclusion-of-women/"><img src="figure/bechdel.png" width=500></a> --- ## Practice Produce appropriate 5NG with R package & data set in [ ], e.g., [`nycflights13::weather`] <!-- Try to look through the help documentation/Google to improve your plots --> 1. How does `age` predict `recline_rude`? <br> [`fivethirtyeight::flying`] 2. Distribution of log base 10 scale of `budget_2013` <br> [`fivethirtyeight::bechdel`] 3. How does `budget` predict `rating`? <br> [`ggplot2movies::movies`] --- ### HINTS <!-- --> --- class: inverse, center, middle # DEMO in RStudio --- class: center, middle ### Determining the appropriate plot <a href="https://coggle.it/diagram/V_G2gzukTDoQ-aZt"><img src="figure/viz_mindmap.png" style="width: 385px;"/></a> --- layout: false class: center, middle name: tidying # Data Tidying <center> <a href="https://tidyr.tidyverse.org"> <img src="img/tidyr_hex.png" style = "width: 380px;"/> </a> </center> --- layout: true class: inverse .footer[Slides available at http://bit.ly/ness-infer       [Return to Table of Contents](#toc)] --- # Tidy Data? <img src="http://garrettgman.github.io/images/tidy-1.png" alt="Drawing" style="width: 750px;"/> 1. Each variable forms a column. 2. Each observation forms a row. 3. Each type of observational unit forms a table. The third point means we don't mix apples and oranges. --- ## What is Tidy Data? 1. Each observation forms a row. In other words, each row corresponds to a single instance of an <u>observational unit</u> 1. Each variable forms a column: + Some variables may be used to identify the <u>observational units</u>. + For organizational purposes, it's generally better to put these in the left-hand columns 1. Each type of observational unit forms a table. --- ## Differentiating between <u>neat</u> data and <u>tidy</u> data - Colloquially, they mean the same thing - But in our context, one is a subset of the other. <br> <u>Neat</u> data is - easy to look at, - organized nicely, and - in table form. -- <u>Tidy</u> data is neat but also abides by a set of three rules. --- class: center, middle <a href="figure/lebowski-abides-o.gif"><img src="http://stream1.gifsoup.com/view8/20150404/5192859/lebowski-abides-o.gif" style="width: 450px;"/></a> <img src="figure/tidy-1.png" alt="Drawing" style="width: 750px;"/> --- ## Is this tidy? ``` # A tibble: 12 x 4 year title clean_test budget_2013 <int> <chr> <ord> <int> 1 1995 Apollo 13 ok 99370665 2 2005 Brokeback Mountain notalk 16583160 3 2010 Diary of a Wimpy Kid ok 16023478 4 1984 Dune dubious 100864980 5 1984 Ghostbusters notalk 67243320 6 2003 How to Lose a Guy in 10 Days men 63304348 7 2011 Iris ok 5696299 8 2004 Sideways ok 20964279 9 2000 Songcatcher ok 2435235 10 2004 Team America: World Police men 24663858 11 2010 Tron Legacy notalk 213646368 12 2011 War Horse notalk 72498355 ``` --- name: demscore ## How about this? Is this tidy? ``` # A tibble: 12 x 13 country `1952` `1957` `1962` `1967` `1972` `1977` `1982` `1987` <chr> <int> <int> <int> <int> <int> <int> <int> <int> 1 Albania -9 -9 -9 -9 -9 -9 -9 -9 2 Argentina -9 -1 -1 -9 -9 -9 -8 8 3 Armenia -9 -7 -7 -7 -7 -7 -7 -7 4 Australia 10 10 10 10 10 10 10 10 5 Austria 10 10 10 10 10 10 10 10 6 Azerbaijan -9 -7 -7 -7 -7 -7 -7 -7 7 Belarus -9 -7 -7 -7 -7 -7 -7 -7 8 Belgium 10 10 10 10 10 10 10 10 9 Bhutan -10 -10 -10 -10 -10 -10 -10 -10 10 Bolivia -4 -3 -3 -4 -7 -7 8 9 11 Brazil 5 5 5 -9 -9 -4 -3 7 12 Bulgaria -7 -7 -7 -7 -7 -7 -7 -7 # ... with 4 more variables: `1992` <int>, `1997` <int>, # `2002` <int>, `2007` <int> ``` --- name: whytidy ## Why is tidy data important? - Think about trying to plot democracy score across years in the simplest way possible with the data on the [previous slide](#demscore). -- - It would be much easier if the data looked like what follows instead so we could put - `year` on the `x`-axis and - `dem_score` on the `y`-axis. --- ## Tidy is good ```r library(tidyr) dem_score_tidy <- dem_score %>% gather(-country, key = "year", value = "dem_score") %>% mutate(year = as.integer(year)) dem_score_tidy %>% sample_n(10) %>% arrange(country) ``` -- ``` # A tibble: 10 x 3 country year dem_score <chr> <int> <int> 1 Canada 1997 10 2 Costa Rica 1962 10 3 Denmark 2002 10 4 Georgia 1977 -7 5 Montenegro 1972 -7 6 Norway 2007 10 7 Panama 1962 4 8 Sweden 1972 10 9 Taiwan 1952 -8 10 Taiwan 1987 -1 ``` --- ## Let's plot it - Plot the line graph for 4 countries using `ggplot` ```r dem_score4 <- dem_score_tidy %>% filter(country %in% c("Australia", "Pakistan", "Portugal", "Uruguay")) ggplot(data = dem_score4, mapping = aes(x = year, y = dem_score)) + geom_line(mapping = aes(color = country), size = 2) ``` <!-- --> --- ## Beginning steps Frequently the first thing to do when given a dataset is to - check that the data is <u>tidy</u> (if not, convert it!) - identify the observational unit, - specify the variables, and - give the types of variables you are presented with. This will help with - choosing the appropriate plot, - summarizing the data, and - understanding which inferences can be applied. --- layout: false class: center, middle name: sampling # Sampling Distributions <center> <a href="https://moderndive.github.io/moderndive"> <img src="https://raw.githubusercontent.com/moderndive/moderndive/master/images/hex_blue_text.png" style = "width: 380px;"/> </a> </center> --- layout: true class: inverse .footer[Slides available at http://bit.ly/ness-infer       [Return to Table of Contents](#toc)] --- # Review - Write the code to produce this plot for 2007 data 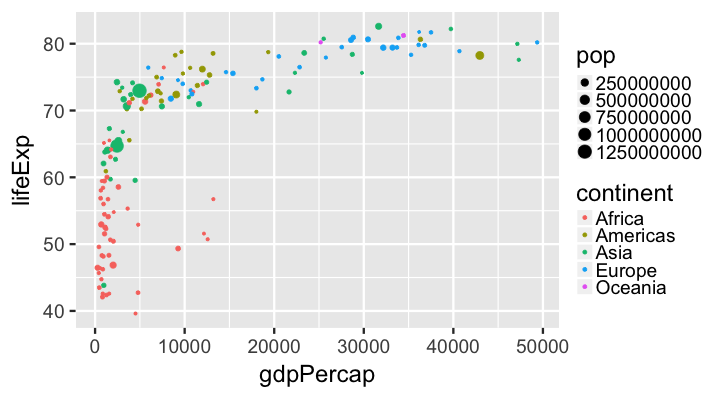<!-- --> --- ## Extending this knowledge to something new - How can we now learn about <u>sampling distributions</u>? -- <center> <a href="http://moderndive.com/images/sampling_bowl.jpeg"> <img src="img/sampling_bowl.jpeg" style = "width: 380px;"/> </a> </center> --- ```r library(moderndive) bowl ``` ``` # A tibble: 2,400 x 2 ball_ID color <int> <chr> 1 1 white 2 2 white 3 3 white 4 4 red 5 5 white 6 6 white 7 7 red 8 8 white 9 9 red 10 10 white # ... with 2,390 more rows ``` --- ## One virtual scoop of 50 balls (one sample) ```r set.seed(8675309) ( jennys_sample <- bowl %>% sample_n(size = 50) ) ``` ``` # A tibble: 50 x 2 ball_ID color <int> <chr> 1 383 red 2 1148 red 3 1834 white 4 1845 white 5 644 white 6 1612 white 7 2344 red 8 2026 red 9 2050 white 10 1065 white # ... with 40 more rows ``` --- # Proportion that are red ```r jennys_sample %>% summarize(prop_red = mean(color == "red")) %>% pull() ``` ``` [1] 0.38 ``` -- ## Is this how many are in the full bowl? --- ## Sampling variability ### What does `rep_bowl_samples` look like? ```r library(moderndive) rep_bowl_samples <- bowl %>% rep_sample_n(size = 50, reps = 10000) ``` -- ### How about `bowl_props`? ```r bowl_props <- rep_bowl_samples %>% group_by(replicate) %>% summarize(prop_red = mean(color == "red")) ``` --- ## The sampling distribution ```r ggplot(data = bowl_props, mapping = aes(x = prop_red)) + geom_histogram(binwidth = 0.02, color = "white") ``` 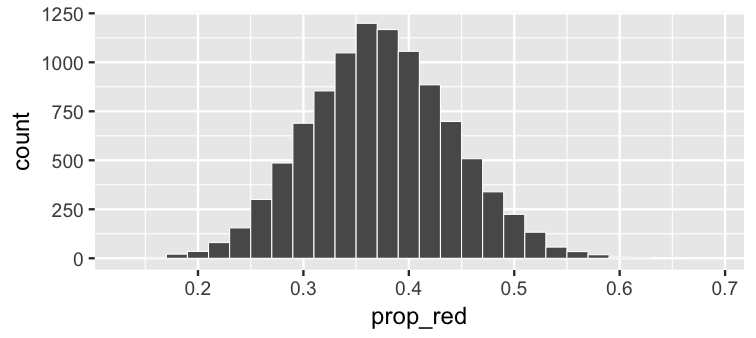<!-- --> --- # Practice - Let's estimate the mean age of pennies - Create a sampling distribution of 10,000 samples each of size 100 from `moderndive::pennies` --- ## Shifting focus ### What about if all we had was the one sample of balls (not the whole bowl)? ```r jennys_sample %>% count(color) ``` ``` # A tibble: 2 x 2 color n <chr> <int> 1 red 19 2 white 31 ``` -- ### How could we use this sample to make a guess about the sampling variability from other samples? --- layout: true class: inverse --- # Building up to statistical inference! ```r library(infer) jennys_sample %>% specify(formula = color ~ NULL, success = "red") ``` ``` Response: color (factor) # A tibble: 50 x 1 color <fct> 1 red 2 red 3 white 4 white 5 white 6 white 7 red 8 red 9 white 10 white # ... with 40 more rows ``` --- # Bootstrapping? ```r library(infer) ( bootstrap_samples <- jennys_sample %>% specify(formula = color ~ NULL, success = "red") %>% generate(reps = 48, type = "bootstrap") ) ``` ``` Response: color (factor) # A tibble: 2,400 x 2 # Groups: replicate [48] replicate color <int> <fct> 1 1 red 2 1 white 3 1 white 4 1 red 5 1 white 6 1 white 7 1 red 8 1 red 9 1 red 10 1 red # ... with 2,390 more rows ``` --- # What does `bootstrap_samples` represent? ## Remember we assumed that all we had was the original sample of 19 red and 31 white to start. -- ## Hope `bootstrap_samples` is close to this: <center> <a href="http://moderndive.netlify.com/images/sampling_bowl.jpeg"> <img src="img/big_sampling_bowl.jpeg" style = "width: 500px;"/> </a> </center> --- ## Bootstrap statistics ```r jennys_sample %>% specify(formula = color ~ NULL, success = "red") %>% generate(reps = 48, type = "bootstrap") %>% calculate(stat = "prop") ``` ``` Response: color (factor) # A tibble: 48 x 2 replicate stat <int> <dbl> 1 1 0.360 2 2 0.320 3 3 0.420 4 4 0.380 5 5 0.540 6 6 0.260 7 7 0.380 8 8 0.480 9 9 0.340 10 10 0.360 # ... with 38 more rows ``` --- ## Do 10,000 reps to get a better sense for variability ### Just as we did with the sampling distribution ```r bootstrap_stats <- jennys_sample %>% specify(formula = color ~ NULL, success = "red") %>% generate(reps = 10000, type = "bootstrap") %>% calculate(stat = "prop") ``` --- ### The bootstrap distribution 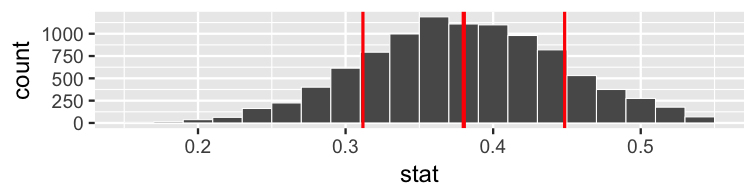<!-- --> ### The sampling distribution 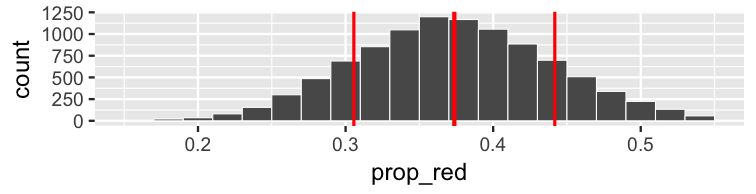<!-- --> --- layout: false ## `infer` verbs <center> <a href="https://infer.netlify.com"> <img src="img/infer_ci.jpg" style = "width: 750px;"/> </a> </center> --- ## `infer` verbs <center> <a href="https://infer.netlify.com"> <img src="img/infer_ht.jpg" style = "width: 750px;"/> </a> </center> --- layout: true class: inverse .footer[Slides available at http://bit.ly/ness-infer       [Return to Table of Contents](#toc)] --- layout: false class: middle, center # Statistical Inference <center> <a href="https://infer-dev.netlify.com"> <img src="img/infer_gnome.png" style = "width: 400px;"/> </a> </center> <br> .footnote[`infer` hex sticker designs kindly created by [Thomas Mock](https://www.linkedin.com/in/jthomasmock/)] --- layout: true class: inverse .footer[Slides available at http://bit.ly/ness-infer       [Return to Table of Contents](#toc)] --- # Research Question If you see someone else yawn, are you more likely to yawn? In an episode of the show *Mythbusters*, they tested the myth that yawning is contagious. - Analysis done with [Alison Hill](https://alison.rbind.io) --- ## Participants and Procedure -- - 50 adults who thought they were being considered for an appearance on the show. -- - Each participant was interviewed individually by a show recruiter ("confederate") who either yawned or did not. -- - Participants then sat by themselves in a large van and were asked to wait. -- - While in the van, the Mythbusters watched to see if the unaware participants yawned. --- ## Data - 34 saw the confederate yawn ( *seed* ) - 16 did not see the confederate yawn ( *control* ) - `1` corresponds to yawn, `0` to no yawn -- ```r group <- c(rep("control", 12), rep("seed", 24), rep("control", 4), rep("seed", 10)) yawn <- c(rep(0, 36), rep(1, 14)) yawn_myth <- data_frame(subj = seq(1, 50), group, yawn) %>% mutate(yawn = factor(yawn)) slice(yawn_myth, c(5, 17, 37, 49)) ``` ``` # A tibble: 4 x 3 subj group yawn <int> <chr> <fct> 1 5 control 0 2 17 seed 0 3 37 control 1 4 49 seed 1 ``` --- # Results <center> <a href="https://github.com/sfirke/janitor"> <img src="img/janitor_hex.png" style = "width: 180px;"/> </a> </center> ```r library(janitor) yawn_myth %>% tabyl(group, yawn) %>% adorn_percentages() %>% adorn_pct_formatting() %>% adorn_ns() ``` ``` group 0 1 control 75.0% (12) 25.0% (4) seed 70.6% (24) 29.4% (10) ``` --- class: inverse, middle, center ## Conclusion -- ## *Finding: CONFIRMED* <sup>1</sup> .footnote[ [1] http://www.discovery.com/tv-shows/mythbusters/mythbusters-database/yawning-contagious/] --- # Really? > "Though that's not an enormous increase, since they tested 50 people in the field, the gap was still wide enough for the MythBusters to confirm that yawning is indeed contagious." <sup>1</sup> .footnote[ [1] http://www.discovery.com/tv-shows/mythbusters/mythbusters-database/yawning-contagious/] --- # State the hypotheses -- Null hypothesis: > There is no difference between the seed and control groups in the proportion of people who yawned. -- Alternative hypothesis (directional): > More people (relatively) yawned in the seed group than in the control group. --- # Test the hypothesis Which type of hypothesis test would you conduct here? - Independent samples t-test - Two proportion test - Chi-square Goodness of Fit - Analysis of Variance --- class: center, middle # Two proportion test -- `\(H_0: p_{seed} - p_{control} = 0\)` -- `\(H_A: p_{seed} - p_{control} > 0\)` --- # The observed difference ```r yawn_myth %>% group_by(group) %>% summarize(prop = mean(yawn == 1)) ``` ``` # A tibble: 2 x 2 group prop <chr> <dbl> 1 control 0.250 2 seed 0.294 ``` -- ```r (obs_diff <- yawn_myth %>% group_by(group) %>% summarize(prop = mean(yawn == 1)) %>% summarize(diff(prop)) %>% pull()) ``` ``` [1] 0.044118 ``` --- class: inverse, middle, center ## Is this difference *meaningful*? -- ## Different question: -- ## Is this difference *significant*? --- # Modeling the null hypothesis If... <br>    `\(H_0: p_{seed} = p_{control}\)`    <br> is true, then whether or not the participant saw someone else yawn does not matter. -- <br> In other words, there is no association between exposure and yawning. --- class: inverse, center, middle  --- .pull-left[ ### Original universe ``` # A tibble: 12 x 3 subj group yawn <int> <chr> <fct> 1 1 control 0 2 2 control 0 3 3 control 0 4 4 control 0 5 5 control 0 6 6 control 0 7 15 seed 0 8 16 seed 0 9 17 seed 0 10 18 seed 0 11 19 seed 0 12 20 seed 0 ``` ``` group 0 1 Total control 12 4 16 seed 24 10 34 Total 36 14 50 ``` ] -- .pull-right[ ### Parallel universe ``` # A tibble: 12 x 3 subj group alt_yawn <int> <fct> <fct> 1 1 control 0 2 2 control 0 3 3 control 0 4 4 control 0 5 5 control 0 6 6 control 0 7 15 seed 0 8 16 seed 1 9 17 seed 1 10 18 seed 0 11 19 seed 0 12 20 seed 1 ``` ``` group 0 1 Total control 14 2 16 seed 22 12 34 Total 36 14 50 ``` ] --- # 1000 parallel universes .pull-left[ ``` Response: yawn (factor) Explanatory: group (factor) Null Hypothesis: independence # A tibble: 1,000 x 2 replicate stat <int> <dbl> 1 1 0.0441 2 2 0.0441 3 3 -0.0478 4 4 0.136 5 5 0.0441 6 6 -0.0478 7 7 0.136 8 8 -0.232 9 9 -0.0478 10 10 -0.0478 # ... with 990 more rows ``` ] -- .pull-right[ ``` # A tibble: 10 x 2 replicate stat <int> <dbl> 1 991 0.0441 2 992 -0.0478 3 993 0.136 4 994 -0.232 5 995 -0.140 6 996 -0.140 7 997 0.136 8 998 0.0441 9 999 0.136 10 1000 -0.324 ``` ] --- ## The parallel universe distribution <!-- --> The distribution of 1000 differences in proportions, if the null hypothesis were *true* and yawning was not contagious. --- ## Calculating the p-value In how many of our "parallel universes" is the difference as big or bigger than the one we observed (0.04412)? -- <br> The shaded proportion is the p-value! ``` Response: yawn (factor) Explanatory: group (factor) Null Hypothesis: independence # A tibble: 1 x 3 n_as_big n_total p_value <int> <int> <dbl> 1 505 1000 0.505 ``` --- class: middle, center layout: false # There is Only One Test! [](http://allendowney.blogspot.com/2016/06/there-is-still-only-one-test.html) --- layout: true class: inverse .footer[Slides available at http://bit.ly/ness-infer       [Return to Table of Contents](#toc)] --- layout: false  ---  ---  ---  ---  ---  ---  ---  ---  ---  ---  --- layout: true class: inverse .footer[Slides available at http://bit.ly/ness-infer       [Return to Table of Contents](#toc)] --- # `infer` Five functions: - `specify()` - `hypothesize()` - `generate()` - `calculate()` - `visualize()` --- # `infer` <small> - `specify` the response and explanatory variables (`y ~ x`) - `hypothesize` what the null hypothesis is (here, `independence` of `y` and `x`) - `generate` new samples from parallel universes under the null hypothesis model: - <small>Resample from our original data <u>without replacement</u>, each time shuffling the `group` (`type = "permute"`)</small> - <small>Do this <u>a ton of times</u> (`reps = 1000`)</small> - `calculate` the statistic (`stat = "diff in props"`) for each of the `reps` </small> --- # `infer` example ```r set.seed(8) null_distn <- yawn_myth %>% specify(formula = yawn ~ group, success = "1") %>% hypothesize(null = "independence") %>% generate(reps = 1000, type = "permute") %>% calculate(stat = "diff in props", order = c("seed", "control")) ``` --- # Visualize the null distribution - `visualize` the distribution of the `stat` <br> (here, `diff in props`) ```r null_distn %>% visualize(obs_stat = obs_diff, direction = "right") ``` <!-- --> --- # Classical inference Rely on theory to tell us what the null distribution looks like. ```r yawn_myth %>% specify(yawn ~ group, success = "1") %>% hypothesize(null = "independence") %>% # generate() is not needed since we are not doing simulation calculate(stat = "z", order = c("seed", "control")) %>% visualize(method = "theoretical", obs_stat = obs_z, direction = "right") ``` --- # Classical inference Rely on theory to tell us what the null distribution looks like. ``` Warning: Check to make sure the conditions have been met for the theoretical method. `infer` currently does not check these for you. ``` <!-- --> --- # Resampling vs Classical (`stat = "z"`) ```r yawn_myth %>% specify(yawn ~ group, success = "1") %>% hypothesize(null = "independence") %>% generate(reps = 1000, type = "permute") %>% calculate(stat = "z", order = c("seed", "control")) %>% visualize(method = "both", bins = 10, obs_stat = obs_z, direction = "right") ``` ``` Warning: Check to make sure the conditions have been met for the theoretical method. `infer` currently does not check these for you. ``` <!-- --> --- ## Practice - Read in/prep the [`mazes` data](http://bit.ly/mazes-gist) ```r library(readr) mazes <- read_csv("http://bit.ly/mazes-gist") %>% janitor::clean_names() %>% filter(dx %in% c("ASD", "TD")) ``` - Use `infer` to compare a numerical variable between the two groups using: - A permutation test and - A classical theoretical test. About the data: <small>[Quantitative analysis of disfluency in children with autism spectrum disorder or language impairment](http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0173936)</small> --- # More practice (time permitting) - Find a data set / use your own and perform statistical inference on one or two of the variables there using the `infer` package --- ## More info and resources - https://infer.netlify.com (https://infer-dev.netlify.com for development) - Many examples under Articles with more to come - To be discussed in [www.ModernDive.com](www.ModernDive.com) - [Sign up](http://eepurl.com/cBkItf) to the mailing list for updates - DataCamp courses that use `infer` - [Inference for Numerical Data](https://www.datacamp.com/courses/inference-for-numerical-data) - [Inference for Regression](https://www.datacamp.com/courses/inference-for-linear-regression) - Two more DataCamp courses to launch soon - [Learn the Tidyverse](https://www.tidyverse.org/learn/) --- ## Future plans - Generalize `calculate()`'s `stat` argument - Improve speed and memory used by shifting to list-columns - Rewrite the inference part of the [ModernDive](http://moderndive.netlify.com/9-ci.html) book - Lead more workshops on using the `infer` package to extend knowledge of the `tidyverse`, R, and statistics --- layout: false class: inverse, middle <center> <a href="https://www.datacamp.com"> <img src="img/DataCamp_Vertical_White.png" style="width: 220px;"/> </a>  <a href="https://moderndive.netlify.com"> <img src="img/dark_vert.png" style="width: 210px;"/></a>  <a href="https://infer.netlify.com"> <img src="img/infer_oregon3.png" style="width: 200px;"/></a></center> ## Thanks for attending! Contact me: [Email](mailto:chester@datacamp.com) or [Twitter](https://twitter.com/old_man_chester) - Special thanks to - [Albert Y. Kim](https://twitter.com/rudeboybert) - [Andrew Bray](https://andrewpbray.github.io) - [Alison Hill](https://twitter.com/apreshill) - Slides created via the R package [xaringan](https://github.com/yihui/xaringan) by Yihui Xie - Slides' source code at <https://github.com/ismayc/talks/>